Through genomics, healthcare is undergoing a revolutionary transformation, providing improved diagnostic tools alongside tailored treatment options and disease prevention strategies. By utilizing genomic sequencing, doctors can detect genetic mutations that lead to diseases, resulting in better and earlier diagnostic outcomes, such as identifying cancer-related mutations or rare genetic abnormalities. The genomic profile of patients informs personalized medicine by guiding treatment selection that targets specific tumor genetic variants. Genomic screening is crucial in preventive medicine by revealing genetic predispositions to diseases, empowering patients to proactively monitor their health and make lifestyle changes.
Redefine Your IT Strategy with Our Consultancy
Customized Solutions for Optimal Performance
Discover IT ConsultingChallenges in Genomics Data Storage and Processing
Genomics presents significant challenges due to its vast volume of data and computational demands. Modern DNA sequencing equipment generates enormous quantities of data, with each human genome raw sequence requiring over 100 GB of storage, while large genomic projects process thousands of genomes. Analyzing 220 million human genomes annually produces 40 exabytes of data, surpassing YouTube’s yearly data output. This vast amount of data necessitates careful management, as it must be secured while allowing easy access over extended periods; traditional on-premise storage lacks scalability benefits. Processing genomic data involves a similar level of complexity.
DNA read analysis requires multiple sequential bioinformatics operations, starting with alignment, followed by variant calling and annotation, all of which depend on robust computing systems and specialized software. Ensuring the reliable and timely execution of these pipelines by researchers and clinicians poses a challenge, making it costly and difficult to maintain in-house high-performance computing (HPC) infrastructure. Genomics greatly benefits from cloud-based solutions due to its substantial data handling needs, complex workflows, and high accuracy and speed requirements.
Cloud-based Solutions for Genomics
To succeed, every business team working with genetic information must utilize optimized cloud architecture solutions. A well-designed solution reduces data processing times, leading to quicker diagnoses and therapy selections for clinical applications. Biotech and pharma organizations benefit from accelerated analysis by shortening their research and development durations and the periods for commercializing new therapies. A properly designed cloud pipeline shifts server management responsibilities to the cloud provider, allowing your team to focus on scientific development rather than infrastructure maintenance. The solution offers dynamic scalability that enables users to process data at scale while reducing costs by scaling down operations.
AWS provides practical solutions to various organizations, enabling them to expedite the conversion of raw sequencing data into valuable insights through their secure, scalable, and economical systems. Genomic organizations of all sizes, from biotech startups to national genome programs, leverage AWS to gain faster insights while decreasing expenditures and upholding data security measures. The following sections outline methods to build a resilient genomics data pipeline on AWS, explaining architectural elements alongside suitable AWS services and proven best practices for delivering technical excellence and business value outcomes.
Architectural Considerations
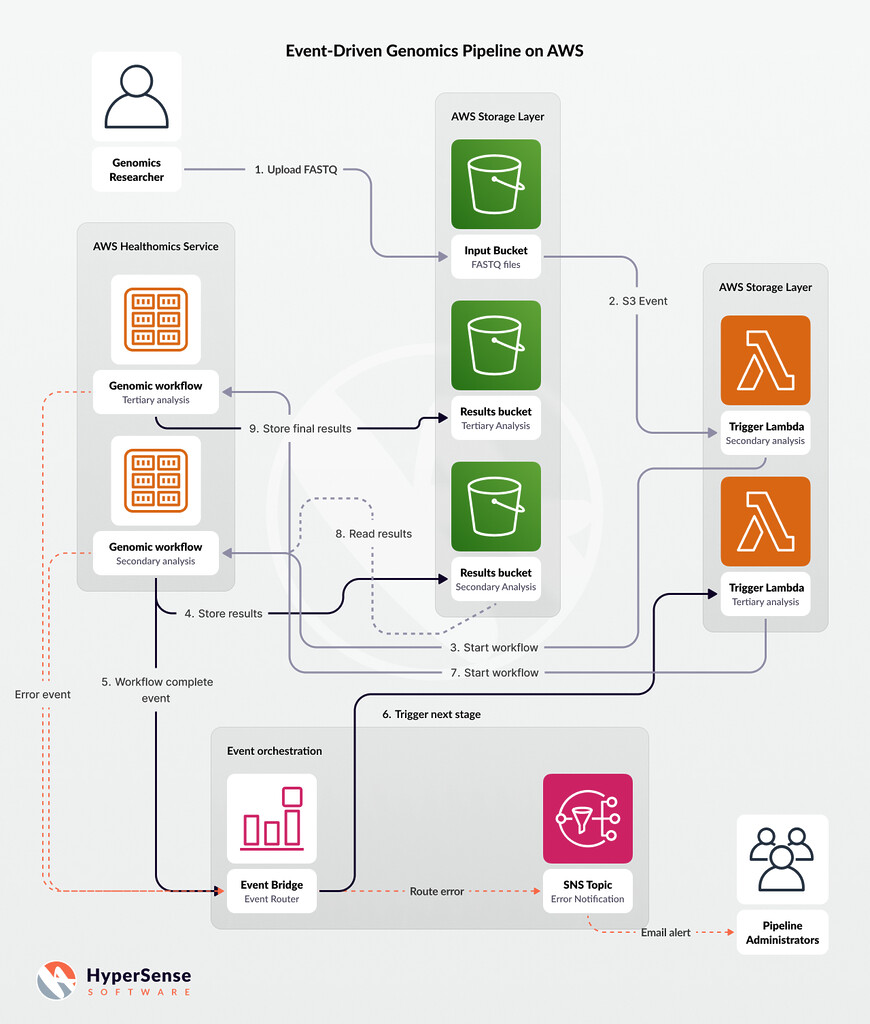
AWS users designing genomics pipelines must develop efficient data flow handling architecture patterns. Event-driven architecture acts as a fundamental solution that is optimal for processing genomic data. In event-driven architectures, system components automatically trigger their processes based on real-time events rather than depending on predefined schedules or human intervention. This pattern creates independent stages in the pipeline that immediately respond to the outcomes generated by other stages. The sequencer uploads raw genome files to storage, which subsequently sparks the analysis pipeline.
AWS services produce native events that act as triggers to commence downstream processing after file storage or queue message events occur. Event-driven pipelines function autonomously and are highly automated, eliminating the need for human involvement between processing stages. This system enhances reliability and throughput, as it begins each process immediately once its prerequisite conditions are met. The genomic process completes faster with event-driven pipelines, as batch jobs do not require manual initiation, and the automated system minimizes human-induced errors. A responsive bioinformatics workflow is founded on events from various services, as detailed by AWS through their event-driven architecture.
Orchestrating Multi-Step Analysis Pipelines
The orchestration of multi-step analysis pipelines typical in genomics operations requires careful attention. A standard bioinformatics workflow involves consecutive dependent tasks, beginning with primary data processing, followed by secondary analysis (alignment and variant calling), and culminating in tertiary analysis (annotation and interpretation). Cloud architecture necessitates configuring various step execution processes and data transfer methods between these processes. Organizations can implement their workflows using AWS native workflow services alongside event buses. AWS Step Functions allow users to manage coordinated sequences of Lambda functions or container tasks through defined state transitions.
AWS EventBridge serves as a serverless event bus that provides an alternative method to manage event routing between different services. A robust pipeline implementation utilizes Amazon S3 events together with AWS Lambda functions and EventBridge rules to activate the following workflow: a new S3 object (such as a raw FASTQ file) triggers Lambda to initiate analysis, and when the analysis is complete, EventBridge executes the next pipeline step. Event chaining facilitates proper stage execution while maintaining loose service relationships. The system supports parallel execution by handling multiple samples simultaneously and offers strong error management capabilities that trigger notifications or corrective actions.
Redefine Your Project with Our Development Teams
Fuel Your Projects with Tailored Software Development Expertise
Get Your Development TeamDesigning a Scalable and Modular Genomics Architecture on AWS
Architectural design must incorporate two key aspects of genomic data characteristics and their large scale. Genomics solutions on AWS depend on four main components: scalable storage systems, processing capacity, messaging systems, and data analysis visualization tools. The central element focuses on data storage for large datasets because Amazon S3 allows users to store petabytes of genomic data with high durability. The elastic compute services from AWS provide processing power for data-intensive procedures, including alignment and variant calling applications via Lambda, Fargate/EKS, and EC2, while AWS Batch supports HPC workloads.
The components of this system integrate through orchestration tools (such as Amazon EventBridge, AWS Step Functions, Amazon SQS, SNS, etc.) that ensure loose coupling among the elements. AWS offers diverse processing services that enable users to create cohesive genomics solutions from end to end. Architectural design involves selecting serverless and managed services to eliminate maintenance tasks (including server setup) by adopting specialized AWS HealthOmics instead of building custom HPC clusters.
A centralized data lake serving raw and processed data should be established to allow different teams and tools to retrieve data for analytical and visualization purposes. A genomics platform architecture built on AWS must adhere to an event-driven design that provides scalability and modularity to ingest large files and operate complex pipelines rapidly before delivering results to researchers or application systems. The architecture must also utilize AWS-managed services for maximum speed, security, and reliability.
AWS Technologies for Genomics
AWS offers numerous services that can be combined to create a comprehensive genomics data platform. Key technologies and services include purpose-built genomics services, general cloud storage and computing, data analytics tools, and AI services. Further, we explore some of the most relevant AWS offerings for genomics and how they fit into a data storage and processing pipeline.
AWS HealthOmics for Genomic Data Processing
The AWS HealthOmics service represents a purpose-built solution for omics data analysis and management that recently became available to customers. This solution operates exclusively to process genomic, transcriptomic, and various ‘omics’ data types throughout their entire lifecycle. Through AWS HealthOmics, organizations can manage large omics datasets using query and analytic functions that help generate health-based insights. The managed environment of the service enables users to focus on scientific work instead of worrying about infrastructure management. HealthOmics performs essential research functions quietly in the background, such as schedule management, compute allocation, and workflow retry protocols.
The service enables the execution of custom pipelines developed using standard bioinformatics workflow languages, including Nextflow, WDL (Workflow Description Language), and CWL (Common Workflow Language), allowing users to easily run their own pipelines on AWS. Support for these portable formats is crucial because many researchers currently maintain their pipelines with them. AWS HealthOmics presents two workflow options: private/custom pipelines and Ready 2 Run pipelines, which incorporate analysis pipelines generated by trusted third parties and open-source projects.
AWS users can access ready-to-use workflows, including the Broad Institute’s GATK Best Practices for variant discovery, single-cell RNA-seq processing from the nf-core project, and protein structure prediction with AlphaFold. A genomics team can utilize optimized pipelines through HealthOmics, relying on AWS for setup, tool installation, and environment configuration, which manages workflow containerization on scalable compute resources. The HealthOmics platform connects to other AWS services that provide data storage functionality and monitoring capabilities. AWS HealthOmics offers a ready-to-use solution for processing genomics pipelines, providing users with an accessible method to execute complex analyses at all scales.
Amazon S3 for Large-Scale Genomic Data Storage
Amazon Simple Storage Service (Amazon S3) is the essential platform for genomics applications, offering scalable storage with high durability and cost-effective solutions. Amazon S3 enables virtually unlimited file storage capacity of any size, meeting the requirements for storing petabytes of genomic data sets. The durability level of S3 reaches 11 9’s (99.999999999%), ensuring that data stored in S3 is at negligible risk of loss. Preserving valuable genomic datasets necessitates data storage across multiple facilities, along with a high level of durability. Data access in S3 supports parallel usage by multiple users or processes, enabling collaborative research efforts and fast pipeline processing. AWS endorses S3 as a genomics platform because it provides “industry-leading scalability, data availability, security, and performance.”
Data Storage in S3 buckets
Data storage in S3 buckets follows a logical structure that separates project data from data types through FASTQ files, BAM/CRAM files, VCF files, and analysis outputs.
Storage expenses can be minimized through S3’s multiple storage level system, which benefits genomics operations. Standard S3 storage maintains low-latency access to frequently used data, whether during active projects or for recently sequenced samples. The selection of storage class on Amazon S3 depends on data age and usage frequency, allowing costs to decrease as data ages or becomes less frequently accessed post-analysis or when archiving raw data from older studies. Genomics datasets can be stored inexpensively using low-cost options within Amazon S3 Glacier tiers, including Glacier Instant Retrieval, Glacier Flexible Retrieval, and Glacier Deep Archive.
Glacier Deep Archives Data Storage
The highly affordable Glacier Deep Archive enables long-term data storage, though retrieval of files may take hours. Lifecycle policies facilitate the automatic movement of objects through these storage classes based on age or access patterns, optimizing costs without human input. Storing raw genomics data for long-term reproducibility and future re-analysis becomes efficient in S3, as retrieval access is infrequent. The S3 storage system allows organizations to optimize their costs and response times by keeping crucial information in hot storage while transferring less-needed data to cold storage until retrieval is required. This approach leads to significant savings in ongoing storage expenses.
S3 offers integrated capabilities with AWS services and event-driven functionality. The S3 platform generates native events through Amazon S3 Event Notifications to detect object creation or deletion actions. Event-driven pipelines require this core functionality to enable a Lambda function to process a file immediately after uploading it to the “input” S3 bucket. The event mechanism of S3 acts as the primary activation trigger for serverless workflows. S3 data can be directly accessed by AWS analytics services (Athena, Redshift Spectrum, etc.), allowing queries to be executed within the S3 storage space without moving the data elsewhere. The available encryption and access control features and scalable storage make Amazon S3 the ideal solution for genomics storage, as it manages data costs throughout its lifecycle while enabling other services to access the data reliably.
Amazon Comprehend Medical for Analyzing Clinical Text
Genomic data alone is often an inadequate research tool in many genomics projects where healthcare professionals must integrate medical textual information with genomic findings for meaningful evaluations. The AWS-managed service, Amazon Comprehend Medical, employs natural language processing (NLP) to analyze unstructured medical text, providing valuable support for genomics data processing. Hospital patients can benefit from Amazon Comprehend Medical, which operates as an NLP service that is compliant with HIPAA while detecting and extracting health information from textual documents. The system identifies medical terminology, including diagnoses, medications, procedures, and symptoms, along with Protected Health Information (PHI) within unstructured text. When a clinician’s report enters Comprehend Medical, it extracts specific disease details, gene names, drug names, patient demographic information, and more.
Comprehend Medical Benefits
The genomics pipeline benefits from Comprehend Medical, which connects genomic results with relevant medical information. When genomic analysis reveals genetic variants in a patient, Comprehend Medical enables doctors to extract pertinent phenotypic details from health records and clinical notes related to those specific genetic variants. By extracting information, one can enhance the ability to align DNA variants with clinical traits to understand genetic-to-phenotypic correlations better. Comprehend Medical offers research capabilities for large repositories of scientific publications or clinical trial descriptions, allowing the system to identify gene-outcome associations that improve genomic data interpretation. Through entity recognition and relationship extraction, the service provides AI-enhanced analysis beyond basic data, enabling the identification of drug-condition relationships in medical text. Due to its fully managed nature, developers can access Comprehend Medical’s API for text analysis. They receive structured results that pinpoint notes containing “BRCA1-positive breast cancer, Stage II” diagnosis or mentioning Trastuzumab medication.
The integration of Amazon Comprehend Medical into genomics pipelines streamlines text data processing and ensures compliance by scanning text for PHI while supporting de-identification. Its HIPAA-eligibility status allows healthcare providers to securely incorporate the system into processes that manage patient data. This service enables AWS genomics solutions to analyze both numerical and unstructured clinical information, delivering a more comprehensive analysis. Organizations can develop the complete clinical insights necessary for precision medicine and decision-making by combining genomic data with information extracted from clinical narratives.
Data Lakes and Analytics with AWS Glue, Athena, and Redshift
Once genomic data has been processed (variants identified, results generated), the next challenge is how to aggregate, query, and derive higher-level insights from it – often by combining it with other data sources. AWS provides powerful tools to build a genomics data lake and analytics platform. A typical pattern is to store curated results and metadata in Amazon S3 as a data lake, use AWS Glue to catalog and transform that data, and then use Amazon Athena or Amazon Redshift for interactive querying and analysis.
AWS Glue
AWS Glue provides a fully managed extract-transform-load (ETL) service that is highly beneficial for preparing genomics data. Glue enables automated schema inference from S3 data crawls, resulting in the creation of a Data Catalog metadata repository that contains tables for variant data and sample information. Once added to the catalog, the data becomes searchable using SQL-based engines. Glue’s ETL capabilities allow users to write transformation and enrichment jobs in Python or Scala, which execute on Apache Spark. In genomic applications, Glue is used to convert raw output files into formats suitable for analysis. The text-based Variant Call Format (VCF) files can be transformed into Parquet, a more efficient format for querying needs.
A Glue Spark job can read thousands of VCF files stored in S3 while applying filters or annotations to generate a consolidated Parquet output, partitioned by chromosome or patient. Glue jobs can run through automated pipelines by scheduling them to execute after analysis pipelines complete their operations to update lake-based aggregated data. Glue provides users with the ability to run PHI scans and virus checks on incoming data through its Python shell job features, protecting sensitive data from unauthorized exposure. Organizations can create a governed data lake through Glue’s data orchestration and transformation, while defining genomic outputs as structured datasets that are easily queryable.
Amazon Athena
With data organized and cataloged, Amazon Athena provides a convenient way to run interactive queries on the genomic data lake using standard SQL without setting up any servers. Athena is a serverless query engine that can read data directly from S3 (leveraging the Glue Data Catalog for schema information). Analysts and scientists can write SQL queries to perform tasks such as finding all patients with a specific genetic variant, calculating allele frequencies in a cohort, or joining variant data with phenotype data, all through Athena. Because it is serverless, Athena automatically scales, and you pay per query, making it cost-efficient for ad-hoc analysis.
Amazon Redshift
Amazon Redshift is another key service for managing larger analytics tasks that require complex processing. The Amazon Redshift cloud data warehouse, part of AWS, is designed to handle large volumes of structured data with high-performance capabilities. Redshift can process genomic data in two ways: by storing variant results for all patients in tables for BI dashboards and SQL query support, or by using Redshift Spectrum to perform S3 queries without data loading. With Redshift Spectrum, users can query their S3-based data without needing to create a full copy in the warehouse, thanks to a feature that functions similarly to Athena.
AWS explains that Redshift and Athena serve distinct purposes: Redshift delivers optimal performance for routine queries and dashboard operations, while Athena provides serverless access to datasets with limited traffic. Organizations implement both Athena and Redshift together to address the different operational needs of their customer base. The genomics platform leverages Redshift to power an interactive web portal for patient genomic report retrieval with sub-second response times, whereas data scientists use Athena to query raw research data stored in S3 that is not housed in the warehouse.
The combination of AWS Glue, Athena, and Redshift enables organizations to create genomic data lakes and advanced analytical capabilities. Glue’s data preparation features allow users to catalog their information, while Athena offers exploratory flexibility, and Redshift provides high-performance analytics with BI tool integration. Companies can utilize these components for tertiary genomic analysis to integrate genomic information with clinical data for pattern discovery and build machine learning models via Amazon SageMaker that extract data from the data lake. The architectural design of the data lake transforms processed data from the pipeline into a searchable knowledge repository, empowering organizations to generate business insights through re-analysis.
Scalable Computing for Genomics
Scalability in genomics computing is critical because workloads can range from executing thousands of tiny tasks (e.g., formatting data, and running a small script for each sample) to running a single heavy algorithm that consumes 48 hours of CPU/GPU time. AWS offers several compute paradigms to address this spectrum, primarily serverless functions, serverless containers, and managed Kubernetes.
AWS Lambda for Event-Driven Genomics Pipelines
AWS Lambda acts as the primary serverless function service offered by AWS to its users. This service enables you to execute functions through code triggers, eliminating the need for server maintenance. Lambda serves as a crucial solution for developing glue and trigger systems that support extensive genomic analytical processes. The invocation of Lambda functions aids in processing incoming files and performing simple data operations, such as manifest parsing and sending system alerts. When numerous events trigger Lambda functions, they automatically scale to manage hundreds or thousands of invocations simultaneously, making them ideal for handling file bursts from high-throughput sequencers.
The Lambda service allows for stateless tasks that run continuously for up to 15 minutes, fulfilling multiple auxiliary requirements. The “fan-out” approach with Lambda enables parallel task execution, where one Lambda function triggers N analysis jobs for N samples, followed by another Lambda function aggregating the completed results. There is no need for infrastructure management, and the service provides immediate scalability with pricing based on actual utilization time. Lambda delivers high-cost efficiency for tasks that stay within its allocated runtime and memory limits. AWS genomics workflows frequently use Lambda as an event handler, preventing users from maintaining constant helper servers.
Advanced R&D Solutions Tailored for Your Business Growth
Innovation Is at the Core of Every Product Through R&D-Focused Software Development
Learn About R&D ServicesAWS Fargate and Containerized Processing
When handling analysis tasks that require additional processing power or extended execution times, container-based solutions are the optimal choice. AWS Fargate offers a serverless compute environment that runs Docker containers within Amazon ECS or Amazon EKS. Fargate allows users to run Docker containers as needed without managing the underlying EC2 instance infrastructure. Docker images, along with specialized environments, are beneficial for genomic analysis because many bioinformatics tools and pipelines are available in this format. Fargate enables users to build containers that encapsulate alignment tools and machine learning models, allowing these to be executed at scale in the cloud, with costs based on the duration of container usage.
The service provides the minimum CPU and memory capacity required by the container before automatically shutting it down after execution. Fargate enables simultaneous deployment up to its service limits when you need to execute 100 parallel containers for processing 100 samples. Fargate operates on an elastic model that supports significant processing capabilities whenever needed while freeing users from managing clusters. Fargate performs better than Lambda when handling longer or heavier workloads that require one hour of processing time, 32 GB of memory, or application-specific libraries. The management of containers allows users to utilize any programming language and applications through Fargate, whereas Lambda restricts operations to specific runtimes. The genomic analysis jobs that run in containers are executed through Fargate or AWS Batch (which supports Fargate or EC2) by numerous AWS genomics solutions because these jobs require extended processing times or rely on third-party tools.
Kubernetes on AWS for Orchestrated Workflows
Organizations utilizing Kubernetes platforms can deploy their Kubernetes control plane on AWS using the managed solution known as Amazon Elastic Kubernetes Service (Amazon EKS). With Amazon Elastic Kubernetes Service, users can deploy Kubernetes clusters within AWS while avoiding master node management responsibilities, managing worker nodes independently, or deploying Fargate serverless Kubernetes pods. Genomics teams implement Kubernetes platforms because their existing workflows are already integrated with orchestration systems such as Argo Workflows or Nextflow Tower, which support Kubernetes. The migration of their workflows to the cloud becomes straightforward with EKS, as Kubernetes manages job scheduling, scaling, and container orchestration tasks.
Kubernetes on AWS supports a wide variety of AWS instances, enabling management of GPU-equipped instances for AI/ML processes alongside high-memory instances for genomic assembly applications within a single cluster. The integration between EKS, AWS networking, and IAM provides security and compliance features through IAM roles that govern pod access to AWS services. Kubernetes offers excellent flexibility for any containerized workload, along with custom scheduling and stateful service deployment, but demands more operational management than purely serverless solutions. CTOs and tech leads often combine managed services when possible, such as Lambda, Fargate, and HealthOmics, with EKS or EC2 clusters when they require direct control for specialized HPC tasks and existing containerized pipelines.
A genomics pipeline architecture enables various computing options to function as a unified system. The orchestration function utilizes Lambda, while analysis tasks are executed on AWS Fargate, EKS supports a continuously active supplementary service. AWS provides event-driven and integration capabilities that allow these components to exchange data and messages seamlessly. This solution establishes an architecture that offers scalability, ranging from Lambda-type instant function scaling to container-led large-scale parallel batch processing, thus ensuring consistent performance as data demands grow. Serverless computing and containerization contribute to reducing expenses by making resources available only during productive work periods. AWS offers a comprehensive range of computing solutions that empower users to choose optimal environments for pipeline stages based on speed, memory, concurrency, or cost-effectiveness criteria.
Security, Compliance & Cost Optimization
When handling sensitive biomedical data, such as human genomes and patient health information, security and regulatory compliance are essential factors. Furthermore, genomics pipelines must be designed with cost optimization in mind, considering the scale of data storage and computing required. AWS offers tools and best practices to address both concerns.
Security and Compliance in Genomics Pipelines
The security framework and extensive certifications of AWS empower genomics organizations to meet their legal obligations for HIPAA compliance regarding protected U.S. health information, as well as GDPR requirements for European data protection. The cloud security model mandates that customers protect their data, while AWS secures the infrastructure elements for which it is responsible. AWS facilitates compliant operations through its ready-to-use services and provides security tools for data protection. At the time of this article’s publication, AWS has made over 130 services eligible for HIPAA while supporting various compliance standards, including GDPR and HITRUST. Selecting AWS services for managing personal health data is permitted once a Business Associate Agreement (BAA) is established. AWS’s HIPAA eligibility program encompasses all its services, such as Amazon S3, EC2, Lambda, and Amazon Comprehend Medical, as well as genomics pipeline services. The platform adheres to strict international standards and holds more security certifications than any other cloud provider, ensuring users can trust that the platform meets stringent security benchmarks.
Security and Compliance Best Practices
On the customer side, there are several best practices to ensure the security and compliance of genomics data in AWS:
- Data Encryption: Always encrypt genomic data, both at rest and in transit. S3 can automatically encrypt all objects using either SSE-S3 or SSE-KMS for managed keys. For data in transit, use HTTPS and secure protocols, which AWS services enforce by default for public endpoints. If you are using EBS volumes for EC2 or databases for metadata, enable encryption on those as well. Encryption ensures that even if data is accessed without authorization, it remains unintelligible without the keys.
- Access Control and Identity Management: AWS Identity and Access Management (IAM) should be used to establish the minimum required access privileges for users. AWS IAM provides a way to grant permissions to users, services, and applications that precisely match their necessary requirements. A Lambda processor for genomes should only have read permission for a designated input S3 bucket and write permission for a designated output bucket. EC2 instances, along with Lambda functions, should utilize IAM Roles instead of embedding credentials within the code to ensure the use of managed short-lived credentials. To prevent access issues with production genomic data, consider placing development systems and production systems in separate AWS accounts and implementing resource tagging with IAM condition rules.
- Network Security: The pipeline should include EC2 instances and EKS clusters placed in private subnets of an Amazon VPC to prevent internet exposure. Security groups should control all traffic entering or leaving the system by defining which IP addresses and services can access the EC2 instance serving pipeline steps (basing these rules on bastion host addresses and necessary APIs and S3 buckets). VPC endpoints enable you to access S3 and other services through private endpoints that prevent public internet traffic exposure even within the AWS environment. Network isolation provides additional data protection through its mechanisms.
- Monitoring and Auditing: AWS CloudTrail should enable logging for all API calls as well as resource modifications that occur in your account. The audit log generated by AWS CloudTrail allows users to review access and modification events related to genomic data and infrastructure. The monitoring system based on Amazon CloudWatch should trigger alerts when it detects large data exfiltrations from S3 storage or when access denied errors occur more frequently than expected. These actions may indicate an intruder attempting to explore the system. Among its offerings, AWS provides both Amazon GuardDuty and Amazon Macie; GuardDuty employs log anomaly detection to identify threats, while Macie conducts intelligent S3 bucket scanning for sensitive data. Macie detects personal data and unexpected genomic information stored outside authorized areas, thereby providing notifications for compliance purposes (for instance, when a user uploads medical records to the wrong bucket).
- Compliance Management: HIPAA compliance requires you to establish a signed BAA agreement with AWS. Under GDPR regulations, you should create procedures that enable data subjects to exercise their rights, facilitating data deletion upon request. This necessitates building data catalogs that help identify subject-specific data for deletion. Combining the AWS Glue Data Catalog with tagging or an appropriate data lake structure supports this goal. AWS allows users to define data residency rules by selecting regions where their data must remain, prohibiting its relocation outside the chosen areas, thus adhering to GDPR data localization requirements. Establishing dedicated data lakes in different regions provides a solution for international collaborations aimed at preventing issues related to cross-border data movement.
- Shared Responsibility and Training: Educate your team on AWS security features and the shared responsibility model. Make sure everyone understands that while AWS provides secure building blocks, it is up to the users to use them correctly. Employ the “security by design” principle – incorporate security reviews into your architecture decisions and automate compliance wherever possible (for example, using AWS Config rules to ensure buckets aren’t public or Terraform scripts that enforce encryption settings).
By following these practices, businesses can build genomics pipelines on AWS that safeguard sensitive data and adhere to regulations. Many healthcare and life science organizations have successfully met strict compliance requirements using AWS, leveraging the platform’s built-in capabilities to strengthen security posture. In short, AWS provides the tools to make a genomics solution powerful but also trustworthy and compliant, which is essential when dealing with personal genetic information.
Cost Optimization in Genomics Workloads
Running large-scale genomics workloads in the cloud offers tremendous agility, but effective cost management is essential to ensure the solution remains economically sustainable. Fortunately, AWS’s flexible pricing and services provide numerous ways to optimize data storage and computing costs. Below are some strategies and considerations for cost optimization in an AWS-based genomics pipeline:
Intelligent Storage Tiering
HIPAA compliance necessitates the establishment of a signed BAA agreement with AWS. In accordance with GDPR regulations, you must implement procedures that enable data subjects to exercise their rights by facilitating data deletion upon request. This involves creating data catalogs that assist in locating subject-specific data for removal. By combining AWS Glue Data Catalog with tagging or an appropriate data lake structure, you can achieve this objective. AWS enables users to define data residency rules by selecting regions where their data must remain, thereby preventing its relocation outside the chosen areas and supporting GDPR data localization requirements. Establishing dedicated data lakes in various regions provides a solution for international collaborations that seek to avoid challenges associated with cross-border data movement.
Right-Sizing and Auto-Scaling Compute
Choose EC2 instances and Kubernetes nodes with instance types that align with workload requirements. Memory-optimized instances are necessary for executing in-memory genomic computations, while GPU instances are used solely for GPU-accelerated tasks. Implementing auto-scaling groups and AWS Batch-managed scaling eliminates the need for excessive CPU and memory provisioning to account for hypothetical situations. You only pay for high-power computing resources while actively running jobs.
Streamlining Your Path to Effective Product Discovery
Make Your Ideas a Reality in Four Weeks with Our Results-Driven TechBoost Program
See Product Discovery ServicesUtilize Spot Instances for Batch Workloads
AWS Spot Instances provide users access to EC2’s spare capacity at prices 70-90% lower than standard on-demand rates. Genomic analyses, such as genome alignment and variant calling, act as batch operations that retain their status after interruptions. Your pipelines can withstand faults, and the AWS Batch service can automatically retry interrupted computing jobs, making Spot Instances suitable for most computational tasks. The savings from using Spot Instances become particularly significant when applied to large studies. Utilizing Spot Instances for EC2 instances running alignments and variant calling can greatly cut the cost of processing 1,000 genomes. Support for both on-demand and Spot compute is available in HealthOmics workflows alongside AWS Batch.
Serverless and Managed Services
Select serverless services such as AWS Lambda, Fargate, and Athena whenever possible, as their pricing model does not include idle fees. Serverless architectures provide cost advantages by eliminating persistent server maintenance expenses. Activating Lambda for job execution incurs no charges when inactive, but it does levy minimal rates per operation (just a few pennies per execution). The Athena service from AWS charges based on queries and data scans, making it more economical than maintaining a full database server. AWS offers a variety of services and pricing options, allowing users to develop optimized spending strategies through a “mix and match” functionality, which aligns with AWS’s cost optimization guidelines for both compute and storage.
Monitoring and Cost Governance
AWS provides tools for cost management that enable users to monitor their expenses. Cost analysis through AWS Cost Explorer reveals which pipeline components incur the highest expenses by indicating whether EC2 computing, S3 storage, or data transfer is the primary cost driver. The identified cost factors help you choose specific cost reduction strategies, such as relocating data to more affordable storage when storage costs are high or optimizing code when compute time is the main cost driver. AWS Budgets, equipped with alert settings, allow users to receive notifications when their monthly costs exceed established thresholds. Resource cost allocation should be done by tagging by project or department to illustrate which research projects and products utilize specific resources for better return on investment analysis.
Data Egress and Transfer
Carefully evaluate the expenses associated with data movements. AWS data transfer between services operating in the same region is either free or incurs minimal costs (for example, transferring data from S3 to EC2 within the same region is completely free). Data transfers out of AWS will incur costs when moving data to the internet or to on-premises infrastructure. Providing large datasets to collaborators should involve controlled cloud-based access through either pre-signed S3 URLs or AWS Data Exchange, rather than copying and distributing the data. Transferring large datasets outside the cloud is more economical with AWS Snowball devices in conjunction with Transfer Acceleration, compared to traditional download methods. Data processing costs remain low when you perform most operations within the cloud region where your data is stored.
Optimize Data Formats and Queries
In the analytics phase, storing data in compressed, columnar formats (Parquet/ORC) and partitioning it (by date, genome, etc.) will reduce the amount of data scanned by Athena or Redshift Spectrum, thereby reducing query costs and improving performance. As one AWS whitepaper suggests, sorting and partitioning data can “improve performance and reduce costs” for Athena/Redshift queries. This is a form of cost optimization through efficiency: faster queries mean less compute time billed.
Businesses can maintain their performance requirements by implementing strategies to reduce cloud-related expenses significantly. Organizations running genomics workloads on AWS with optimal optimization can achieve both rapid execution and high scalability while benefiting from lower unit pricing (per sample or terabyte) than traditional methods. AWS regularly introduces new pricing plans, including Savings Plans, for consistent usage and costs; therefore, each time they release more efficient instance types, it makes sense to review your pricing strategy frequently. Optimizing genomics costs on AWS involves selecting appropriate tools that you deactivate when unnecessary, along with AWS pricing models and workflows designed to prevent resource waste. The cloud enables precise resource management because you can easily adjust your resource consumption to discover the best cost-performance balance, which evolves with changing workload requirements.
Implementation Best Practices

Having the right architecture and services is one side of the coin; the other is implementing the solution with best practices to ensure scalability, maintainability, and efficiency. Further, we outline some best practices for developing and operating genomics data pipelines on AWS, touching on technology choices like Node.js, computing models, and workflow management.
Use Node.js for Event-Driven Glue Code
Node.js is a top choice for event-driven application development due to its asynchronous I/O model, which operates without blocking, and its rapid startup time. The AWS platform supports Node.js as a Lambda runtime option, effectively serving as the glue component for a genomics pipeline. A Node.js Lambda function acts as the trigger point for S3 events, allowing access to file metadata before initiating HealthOmics workflows. The swift deployment of Node.js, combined with the efficient operation of its V8 engine, enables your AWS Lambda functions to achieve quick execution times suitable for real-time pipelines. Node.js asynchronous patterns should enhance throughput by performing S3 reads and API calls using either async/await or promise-based methods to prevent function idling.
Node.js offers compact Lambda package sizes by combining only the runtime with your script and several NPM libraries, which decreases cold start durations. It provides complete asynchronous functionality for AWS SDK clients such as S3 and DynamoDB, enabling smooth parallel execution of multiple tasks through event chaining within a single function. Node.js boasts a massive selection of NPM libraries that allow developers to integrate JSON or CSV parsers and genomic data processing capabilities into their Lambda deployments. Additionally, Node.js provides an event-driven system with minimal boilerplate, making it ideal for teams experienced in JavaScript and TypeScript. All business logic within Node.js Lambda functions should remain focused and transactionally independent. Node.js functions should act as reliable orchestrators to handle events in the pipeline while delegating complex or long-running processes to dedicated services or containers.
Serverless vs. Containerized – Choose the Right Compute Paradigm
The implementation decision revolves around choosing between serverless functions and containerized services with long-running servers as viable options. Serverless computing (Lambda) excels at tasks that involve event-driven functionality, short executions, and horizontal scaling capabilities. Suitable serverless tasks include workflow triggering, notification delivery, ETL conversion, and parallelizable operations that can be divided into multiple independent Lambda jobs. This system provides users with three significant advantages: minimal server management, automatic scaling, and request-based payment. The containerized approaches (ECS/Fargate or Kubernetes) represent the best solution for applications requiring a customized runtime environment and longer processing times. You would create a Docker container to run third-party tools because the Lambda runtime does not support these tools or their specific libraries.
Fargate containers alongside EC2 instances offer better solutions than Lambda when a task exceeds 15 minutes of execution duration, thus requiring more time than Lambda’s limit allows. Using containerization proves beneficial for handling the continuous operation of complex services, including databases or web applications that provide API access to platform results. When executing genomic tasks on AWS, developers should combine serverless components for orchestration and lightweight operations with Docker containers (or AWS Batch tasks) for processing. You can leverage the advantages of serverless for cost and management but use containers when additional flexibility becomes necessary. AWS Fargate provides a solution that merges serverless and container operations, allowing users to run containers without managing servers while achieving serverless workload capabilities. Enhance your pipelines’ execution mode switching capabilities by implementing abstraction, which enables an easy transition between local cluster execution and AWS Batch execution.
Experience the Power of Professional Web Development
Transformative Web Solutions Designed for Your Business Growth
Discover Web DevelopmentAutomate Data Ingestion and Workflow Orchestration
The best practice involves pipeline automation, minimizing manual processes in data transfer and analysis initiation. AWS services should be implemented to ensure a continuous data flow between ingestion and results production. The automated transfer of new sequencing data from local NAS storage to S3 can be achieved through AWS DataSync. The event notification system should activate processing once data arrives in S3. AWS Step Functions should be considered as a workflow management solution unless HealthOmics meets specific user needs. AWS Step Functions enables the coordination of Lambda invocations and ECS tasks through a defined error handling procedure that supports multiple retries.
The workflow can become easier to manage by allowing each step to pass its output to the next step as input. Utilizing either WDL or Nextflow workflow languages alongside AWS Batch servers functions as a backend system to manage task queues. Pipeline automation must operate the process automatically so that raw data transforms into analysis results without human intervention at each step. Consistency is achieved through quicker execution times while the process accelerates. The implementation of these workflows should be automated through Code as Infrastructure standards, including CloudFormation or Terraform templates and AWS CDK for both infrastructure components (buckets, functions, Batch queues, etc.) and automated logic (Step Function state machines, etc.). This approach ensures your deployment is repeatable for all development stages and team member handovers.
Accelerate Your Success with Custom Software Development
Propel Your Business Forward with Bespoke Software Solutions
Explore Custom SoftwareManage and Monitor Workflow Execution
Complex pipelines can encounter occasional issues when jobs fail due to bad inputs, particularly when service limits are exceeded. Robust monitoring and logging systems are essential requirements. Each pipeline segment should utilize Amazon CloudWatch Logs for effective logging capabilities, as Lambda functions and other AWS services automatically log to this service. Lambda functions must document their operational steps along with essential IDs, including file names and sample IDs. All events and metrics produced by HealthOmics or Batch should be recorded. Additionally, Amazon CloudWatch Alarms must be configured with Amazon SNS notifications to monitor failure events.
The SNS topic notifies both the engineering team via email or Slack whenever workflow failure events occur, as illustrated in the architectural design. This setup allows team members to gain immediate awareness of problems so they can act swiftly. Adding idempotency along with retry capabilities to your pipeline will enable it to automatically retry failed steps multiple times. Each step within Step Functions allows you to define its own retry procedures. When implementing queuing systems based on SQS, operators can establish dead-letter queues to store messages that fail processing repeatedly, keeping them safe for later evaluation. The process of error detection and execution monitoring enables your pipeline to automatically recover from issues or prevent silent data loss while maintaining system resilience.
Optimize Data Formats and Transfers in Workflows
Pay special attention to the data transfer processes between pipeline stages during implementation. The large file size of genomics datasets (gigabytes) creates efficiency issues when moving these files between steps, as it slows down pipeline execution and increases expenses. Data processing should be conducted directly at the storage location whenever possible or through data streaming. To minimize memory usage, a Lambda function that requires file summary information should utilize S3 range requests instead of loading complete files. A multi-step container workflow running on ECS/EKS can benefit from using Amazon EFS (Elastic File System) or FSx for Lustre as shared file systems, as they provide better performance than re-downloading data from S3 between steps.
S3 multi-part upload and download enables you to effectively utilize network bandwidth options. The compression of intermediate data files is particularly beneficial for BAM files, and text files can take advantage of GZIP compression to reduce transfer times. AWS Glue and custom scripts should process and minimize data prior to more complex analysis by extracting specific genomic regions rather than processing entire genomes repetitively. The implementation optimizations will enhance system performance while lowering costs.
Testing, Versioning, and Documentation
The genomics pipeline code should be maintained as if it were production software. Write tests for significant Lambda functions and scripts using local AWS SAM CLI and event simulation tools from mocking frameworks. Workflows and container images must be versioned to allow the rerunning of past analyses without modification, which is essential for scientific reproducibility. Nextflow users should pin tool container versions with specific GATK image versions while placing pipeline scripts under source control. Future maintainers need documentation that includes AWS architecture diagrams, data flow descriptions, and instruction manuals for handling typical failure situations. The designed solution ensures both operability and transparency for the team amid changes in personnel and requirements.
Organizations that adopt these implementation best practices will establish efficient, robust, and sustainable genomics data processing systems on AWS infrastructure. The system design includes features to execute events without interruptions and supports scalability for high-volume processing while maintaining resistance to system failures and changes in system design over time. The combination of well-implemented practices leads to streamlined operational processes and quicker delivery of outcomes to scientists and clinicians who use the system.
Business Benefits of AWS-Based Genomics Pipelines
A well-designed genomics data storage and processing pipeline on AWS not only delivers technical advantages but also provides significant business value. By utilizing AWS, organizations can speed up their research and healthcare initiatives while managing costs and unlocking new capabilities. Here are some of the key business benefits:
Reduced Time-to-Insight in Research and Healthcare
Cloud-based genomics solutions substantially reduce the time needed to transform raw data into actionable insights. AWS provides elastic computing resources that allow lengthy local server analyses to complete in as little as minutes or hours by distributing tasks across multiple cloud instances. The 100,000 Genomes Project experienced a 99% performance boost from Genomics England after building their platform on AWS infrastructure. The speed of analysis enables researchers and clinicians to obtain results that lead to quicker decision-making.
Clinical applications benefit from this speed, saving lives through fast genome testing of critically ill infants and evaluating tumors for cancer patients seeking appropriate therapies. Organizational responsiveness and competitive advantage arise from rapid insights, allowing pharmaceuticals to recognize drug targets sooner, enabling biotech start-ups to enhance their product development cycles, and permitting healthcare to deliver precise medical interventions without delay. Through cloud computing, the computational speed limit becomes data availability and research inquiries, as IT infrastructure no longer restricts the speed of discovery.
Cost Efficiency and Scalability in Genomics Analysis
The AWS model enables genomics organizations to eliminate costly on-premise compute clusters, as they can scale their infrastructure up or down on demand. Genomics organizations can activate additional resources as needed (such as launching a thousand simultaneous analysis processes) and decrease them after completing work, avoiding unnecessary expenses. By shifting capital investments into operational costs, the expenses directly correspond to project work. Significant cost savings become possible through this approach, mainly benefiting customers with unpredictable workloads and fluctuating computational needs.
The cost optimization tools outlined in the dedicated section enable AWS users to perform genome analyses at a lower cost per sequence compared to traditional setups. The utilization of AWS by leading companies in genomics allows them to reduce their expenses as they manage increasing data volumes. Organizations gain the ability to undertake projects that they previously could not pursue, thanks to scalability. National labs achieve genome-wide population analysis by leveraging elastic cloud computing resources, which they could not carry out without cloud scalability. A business can expand its operations as needed through this flexible system, which ensures that infrastructure limitations will not hinder their growth. Small organizations can attain enhanced resource capabilities through AWS by accessing top-tier computing facilities beyond their initial capital investment, allowing them to compete against established enterprises.
AI-Driven Insights and Real-Time Processing
Organizations achieve real-time processing and advanced genomics analytics through the deployment of AWS services. Serverless computing, along with event-driven pipelines, enables sequencer data analysis immediately after generation, bringing genomics closer to real-time operations. Tracking pathogen genomes in real-time for outbreak response, as well as genetic testing in emergency settings, requires this capability. The native integration of AWS AI and machine learning tools allows genomics data to be processed using AI technology for enhanced analysis. Amazon SageMaker enables users to input various patient records alongside variant data to create predictive machine learning models that forecast disease risks and treatment outcomes. The cloud empowers users to train intricate models on genomic datasets through its immense computing capabilities.
The new generation of AI models, such as DeepVariant for variant calling and AlphaFold for protein folding, now requires significant computing power. However, AWS provides the necessary infrastructure to run these models as needed. Because AWS actively innovates its platform, the required GPUs and TPUs, along with AWS Trainium and Inferentia chips and specialized services, will be made available for new genomics AI tools as they emerge. A company that implements AWS-based pipelines will have the advantage of integrating cutting-edge AI and ML algorithms into its analysis framework. The outcomes of these sophisticated analytics include algorithms that identify disease-causing variants among millions of possibilities in patient data, as well as NLP models that extract drug targets from scientific texts. The capabilities enabled by these systems lead to business differentiation through two avenues: developing unique AI-enabled product features and offering real-time genomic insights as a service, creating additional market opportunities.
Enhance Collaboration and Data Sharing
Cloud-based genomics offers businesses secure collaboration features as a vital competitive advantage. Research and clinical genomics operations often necessitate multi-institutional partnerships to exchange data with their partners, consortia, and customers across their networks. Storing data and pipelines within an AWS environment enables authorized collaborators to analyze data from around the world through a permission-based system that eliminates the need for local storage. The traditional processes of hard drive transportation and VPN setup become unnecessary since data remains accessible via the cloud platform.
The AWS platform enables you to implement three data sharing methods, including cross-account access, Amazon S3 pre-signed URLs, and AWS Lake Formation. Through its advanced security framework, AWS allows data owners to distribute essential information without risking non-compliance. Global companies benefit from a unified cloud platform, as their teams in various locations can access shared cloud-based data repositories, providing everyone with updated information. Eliminating data silos boosts employee productivity and reduces project durations. The NIH Sequence Read Archive (SRA) exemplifies open accessibility on AWS, which facilitates data democratization. Private organizations can replicate this model using AWS to deliver controlled data access to their consortium members or customers.
Enabling Focus on Core Competency and Innovation
By outsourcing infrastructure management to AWS, businesses empower their teams of bioinformaticians, data scientists, and IT staff to focus on activities that generate greater value. Freed from tasks like server configuration, software installation, and storage hardware management, these teams can concentrate on pipeline development, data interpretation, and new feature creation. Through its AWS HealthOmics service, customers can “focus on science and not the infrastructure,” as stated in the description. This transition enables businesses to execute faster development cycles while increasing their capacity to innovate. Implementing AWS solutions allows for the development of new innovative products, alongside enhanced client service and operational adaptability. Organizations can mitigate their risks through AWS cloud deployment, as its expert management ensures reliable operation with a reduced likelihood of infrastructure-induced data loss or system downtime that could incur significant costs.
The implementation of a complete AWS-based genomics platform delivers quick results alongside expandable capabilities and cost savings, which leads to business success. The cloud architecture gives businesses three main advantages: speed to market, cost reduction, and innovative data-driven solutions, which help companies meet their goals.
Future Trends of AWS Genomics Solutions
AWS delivers an expanding genomic platform that helps organizations address their data processing and computational challenges in this field. A well-designed AWS infrastructure that integrates event-triggered data pipelines with scalable resources, analytics capabilities, and enhanced security measures enables businesses to transform genomic information into actionable insights at unprecedented speed and effectiveness. Over the last decade, AWS has committed itself to the life sciences sector by continually developing specialized services, such as AWS HealthOmics, to meet industry requirements.
Cloud support for genomics is expected to show continuous improvement based on current positive trends. Amazon Web Services is adding new features to its platform through user feedback and technological developments, which will offer future users enhanced automated genomics solutions. The AWS HealthOmics platform may experience increased connectivity with other AWS AI services and new data management tools that enhance the processing of large sequence datasets.
Looking ahead, a few future trends stand out in genomics and how cloud solutions will adapt to them:
Explosion of Genomic Data & Multi-omics Integration
Sequencing costs continue to decrease with broader adoption, driving significant growth in generated data. Genomic DNA has become the primary focus, but scientists are showing increased interest in other omics fields such as proteomics, metabolomics, and transcriptomics. Cloud architectures will evolve to analyze individual omics separately, as well as to conduct comprehensive analyses of multiple omics datasets. AWS’s data lake concept offers ideal conditions for managing various data types by keeping them together and linking them, with Glue Data Catalog and Lake Formation serving as examples of data management tools. The future will see additional optimized storage solutions and indexing features from AWS for real-time retrieval of large sequence data across extensive genomic databases.
AI and Machine Learning in Genomics
AI is set to play an even larger role in deriving insights from genetic data, from predicting protein structures (as AlphaFold has done) to identifying regulatory elements in the genome or forecasting disease risks. The future of genomics is intertwined with machine learning and AI, and AWS’s machine learning ecosystem (SageMaker, TensorFlow on AWS, specialized hardware like AWS Trainium chips) will be a crucial asset. We expect AWS to potentially offer pre-built genomics AI models or partner solutions that can be readily applied. Additionally, as generative AI techniques progress, there may be services for simulating genomic variants or generating synthetic genomic data for research. AWS’s support for these computationally intensive tasks ensures that organizations can leverage AI without building supercomputers on-premise.
Real-Time and Edge Genomics
Nanopore sequencers, along with other portable sequencers, bring genomic data generation capabilities closer to the locations where care is provided or needs arise. Edge computing offers a potential solution by performing initial basecalling and analysis on-site before transferring results to cloud services for large-scale operations. Genomic laboratories aiming for on-site computational power with cloud data integration features should consider implementing AWS IoT and Greengrass services. The establishment of real-time analytics pipelines using AWS Kinesis Streams or similar solutions should enable pathogen genome monitoring programs in wastewater and public health air surveillance systems. AWS plans to connect streaming data capabilities with HealthOmics and data lakes for continuous ingestion and analysis of real-time sequencing data.
Enhanced Security and Data Governance
Healthcare institutions will face increasing challenges in protecting patient privacy while facilitating the advancement of genomic research. Advanced data-sharing systems, such as federated analysis, will emerge to enable the analysis of distributed datasets without relocating the data from its storage location (AWS confidential computing and AWS Clean Rooms could assist in implementing this approach for controlled collaborative work). AWS plans to enhance its compliance tools with automated genomic data identification features, similar to how Macie manages PII detection, by providing a future capability to detect and secure human genomic data. Processing genomic data in the future could be improved by the encryption-in-use capabilities that the cloud platform offers through homomorphic encryption or secure enclaves.
Collaborative Platforms and Marketplaces
Research platforms are evolving toward environments that enable scientists to exchange both their data assets and analytical instruments and methods. AWS Data Exchange, along with AWS Marketplace, offers existing solutions to distribute data and algorithms. In the future, institutions will likely establish a marketplace for genomics pipelines that would allow workflows to be shared between accounts on AWS infrastructure. This approach would accelerate the distribution of best practices and innovative analysis methods. The reliable AWS infrastructure creates conditions for shared pipelines to execute consistently across all locations.
Leading the Way with AWS-Based Genomics
Organizations that adopt AWS-based genomics data storage and processing pipelines will lead rapid advancements in this domain. The solution offers dynamic adaptability through its flexible framework as new technologies evolve and needs change. AWS provides scalable infrastructure to business owners and technology leaders seeking a platform to sequence ten thousand genomes or process genomic and electronic health records for millions of patients securely and cost-effectively. The genomic field in cloud computing holds bright prospects because AWS continues to enhance genomic breakthroughs with scalable, innovative solutions. Organizations that establish cloud-native genomics pipelines now will effectively harness future breakthroughs, transforming genomic data into practical health discoveries and scientific advances.
What is an advanced data pipeline in genomics, and how does it work?
An advanced genomics data pipeline is a non-interactive system that accesses large amounts of DNA sequence data in stages such as alignment, variant calling, and annotation. It enhances speed, precision, and uniformity in both clinical and research environments.
Why do genetic testing labs need bioinformatics workflows?
There are bioinformatics processes that enable the labs to transform raw sequencing data into medical knowledge. The workflows automate complicated analyses, facilitate compliance, and minimize turnaround time, which is crucial to diagnostics and personalized medicine.
How much data is generated by a whole-genome sequencing pipeline?
The generation of raw data of a complete genome is 100-200 GB. Big data processes large volumes of data each year, requiring scalable, highly performant storage and compute infrastructure.
Why is cloud computing important for genetic testing and bioinformatics?
Cloud computing provides the scalability, flexibility, and speed needed to handle and store large genomic databases. It removes hardware restrictions and provides a secure, compliant infrastructure for operating automated pipelines.
What are the benefits of using AWS for genomics data processing pipelines?
AWS offers specialized services such as HealthOmics, scalable compute (Lambda, Fargate), and storage via S3. Such tools can help labs automate workflows, reduce costs, accelerate analysis, and comply with regulations such as HIPAA and GDPR.
How do labs protect sensitive genomic data in the cloud?
Cloud services such as AWS offer data encryption, access control (based on identity), private networking, audit logs, and compliance standards (HIPAA, GDPR, etc.) — enabling secure data across all pipeline stages and at a genomic and health scale.
How does AWS help reduce cloud costs for genomics pipelines?
Cost optimization in AWS has also been achieved through tiered storage (S3 + Glacier), batch job spot instances, compute auto-scaling, and serverless services (Lambda and Athena), all billed on a per-use basis.
How much does it cost to run a genomics data pipeline on AWS?
This is because costs are based on usage, and labs can save by paying for it. Economies of scale are achieved by maximizing storage levels, processing with spot instances, and eliminating idle server time with serverless services.
What tech stack do modern bioinformatics and genomics pipelines use?
The modern stacks comprise cloud-native stacks such as AWS Lambda, Fargate, Docker, Kubernetes, Apache Beam, Nextflow, and Snakemake, as well as web technologies such as Node.js, Python, and API layers.
What’s the difference between serverless and container-based computing for genomics?
Serverless (e.g., Lambda) is best for short, event-driven functions with no server management. Jobs that require custom environments (e.g., more resource-intensive or long-lasting) are deployed to containers (e.g., Fargate, EKS).
How does event-driven architecture improve bioinformatics pipeline performance?
The pipelines are event-driven, automatically initiating processing steps (e.g., upon file upload) to minimize delays and human error. They are scaled to run faster and can be used for real-time genomics at clinical or research scales.
Can genetic testing labs process genomic data in real-time?
Yes, current pipelines may commence analysis immediately after sequencing, notably with event workflows and streaming applications. This helps in quick diagnosis in critical care or health emergencies in the population.
What is AWS HealthOmics, and how can labs use it?
AWS HealthOmics is a service for running scalable, customizable genomics workflows. It supports industry-standard languages (Nextflow, WDL), manages infrastructure, and interoperates with other AWS services to fully automate the pipeline.