The Shift to Cloud-Based Genomics
Running Genomic Workloads on AWS is transforming how researchers handle an explosion of genomic data—now doubling faster than Moore’s law. With whole-genome sequencing dipping below $1,000, massive projects are producing petabytes of DNA data that traditional on-premises systems can’t store or process cost-effectively. Cloud platforms—especially Amazon Web Services—let organizations ingest, secure, and analyze this deluge without building new data centers. Public resources such as the NCBI’s SRA already rely on AWS object storage for rapid access, and research teams can spin up hundreds of cloud compute nodes for alignment or variant-calling jobs, then shut them down to save money. The result is on-demand scalability, faster turnaround, and effortless global collaboration for modern genomics projects.
Why Run Genomic Workloads on AWS
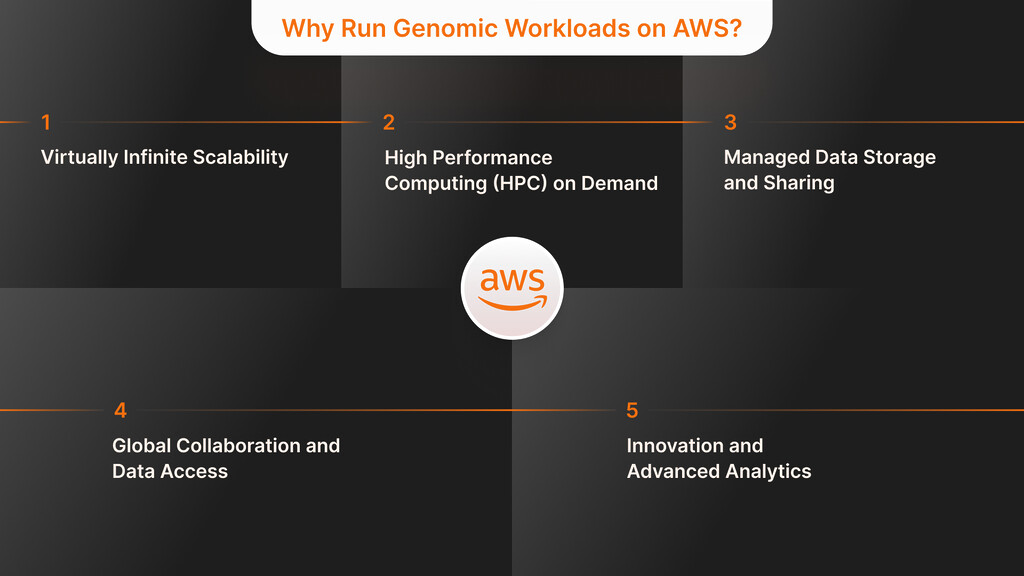
Both technology and business stakeholders find that AWS stands out with its ability to offer larger scale, increased speed, and improved agility for genomic analytics. Genomics workloads involving a lot of data and processing are well supported by AWS’s range of services.
Virtually Infinite Scalability
AWS’s cloud resources can easily manage data bursts in sequencing workloads that are too much for most on-site systems. You can start up many compute nodes whenever you need to run genomic pipelines in parallel. For instance, launching a few hundred AWS instances allowed a research team to analyze their data in just one day, whereas it previously took them weeks, giving scientists space to ask questions they couldn’t before. Performing jobs such as whole genome sequencing alignment and variant calling on dozens or thousands of cores using the cloud is simple. After the analysis, you may turn off services to prevent further expenses.
High Performance Computing (HPC) on Demand
High-performance AWS instances are designed to manage big data and use advanced components, plus network services enhanced for HPC tasks. As a result, even sophisticated genomic algorithms (like genome assembly or secondary analysis) can be used with no significant issues. With AWS ParallelCluster, you can launch parallel clusters with schedulers in the AWS cloud, and AWS Batch is a managed service for running many genomic jobs separately on elastic resources. This means that cloud HPC eliminates the delay caused by queues on shared local clusters, so analysts get their data sooner and can advance R&D more quickly.
Custom Software Development for a Competitive Edge
Build Unique Software Solutions with Our Expertise
Explore Custom SoftwareManaged Data Storage and Sharing
Genomic data is often so large that it takes terabytes to store, and AWS storage is built for safety, high transfer speeds, and accessibility from anywhere. Amazon Simple Storage Service (S3) can store all your sequencing reads, reference genomes, and analysis results in one place. S3 is able to support 11 nines of durability and allows for quick sharing among collaborators, using detailed access rules. Fortunately, tools like AWS DataSync help securely move large amounts of sequencing data from on-site systems or storage to the cloud with little extra effort. In S3, once your data is stored, you may tier it to economical storage or put it in Amazon S3 Glacier Deep Archive for cold data savings. Basically, AWS provides the needed storage scaling, which means you won’t run out of space for your expanding FASTQ files.
Global Collaboration and Data Access
Because the AWS Cloud is global, genomics teams from different parts of the world can use the same resources and data. Researchers can now access each other’s genomic data through the cloud, without shipping hard drives. Many public genomics datasets, such as the 1000 Genomes Project and gnomAD, are already available on open S3 buckets through AWS. You don’t have to download and save them yourself to analyze. The closer the data is to the processing, which is sometimes called reducing data gravity, greatly supports data-intensive science.
Innovation and Advanced Analytics
Thanks to AWS’s wider network, genomics can now be paired with big data analytics and machine learning. Should you need to, you can query your variant call files or expression matrices in S3 by using Amazon Athena or the results in Amazon SageMaker to make predictions. Amazon Omics was launched by AWS as a service that helps manage and query omics data and facilitates large-scale genomic analysis. You can automatically upload genomic data to managed sequence stores and variant stores with Amazon Omics and run workflows (written in WDL or Nextflow) without arranging the necessary infrastructure. AWS manages everything from infrastructure to providing services for life sciences innovation.
To explore AWS-specific tools for managing genomics data, visit Leveraging AWS for Scalable Genomics Data Storage and Processing.
Building Blocks: Key AWS Services for Bioinformatics Pipelines on AWS

Effective use of bioinformatics pipelines in AWS requires selecting the proper services for your genomics toolkit. Cloud genomics workflows make extensive use of these AWS services and tools:
Amazon S3 for Data Storage
As we already explained, S3 is the base platform for managing genomics data on the cloud. This service is excellent for storing initial sequencing data, work in progress, and final results. It grows to practically any size and offers ways to manage old data by moving it to the Glacier archive and replicating data across regions when required. Several genomics pipelines (such as GATK workflows) can take data directly from S3. Good performance depends on your pipeline’s ability to read and write data to S3 without issues.
AWS DataSync and Snow Family for Data Ingestion
Moving big data into the cloud can be tricky. Moving data from your NAS or local storage to AWS is made simpler and faster by AWS DataSync, which provides encryption and verifies it automatically. It can only send new or updated files, which is perfect for keeping up with new sequencing experiments. When you have too much data or a slow internet connection, AWS provides Snowball or Snowmobile devices to ship your data to the cloud. You want to ensure that sequencing data is transferred into your AWS environment as soon as it is produced, which helps your analysis run smoothly.
Amazon EC2 and AWS Batch for Compute
AWS offers different computing choices for major tasks like matching reads, spotting variants, and analyzing the genome. Amazon EC2 enables you to tailor the CPU, RAM, and GPU on virtual machines to suit specific tools, including ones used for genome assembly. In most genomic applications, AWS Batch is highly valuable, as it automatically sets up EC2 instances, queues the jobs, and scales your compute power according to how much work is needed. It is easy to submit your jobs (for example, request 50 instances to run your genome alignment container), and AWS Batch will find the best instances, launch the jobs isolated from one another, and shut them down once the job is done. It allows you to run many batch jobs without worrying about the ops needed to manage them. AWS Lambda (serverless functions) is useful for simple tasks, including starting workflows when new files appear in S3. Still, most genomic computation is carried out on EC2 or Batch for their computing power.
Workflow Orchestration Tools (AWS Step Functions, Amazon Genomics CLI, and more)
Most genomic analyses are carried out in a series of involved stages. Workflow engines offer an option to automate every action in the process. AWS Step Functions is a serverless orchestration service that helps you create state machines that control each part of your pipeline (e.g., start alignment job, then call variants, and finally generate a report). You can have Step Functions call AWS Batch and then wait until the job is done, making it easy to implement a managed pipeline that can retry and run branches simultaneously. In reality, AWS offers reference architectures with Step Functions scheduled for each task, and AWS Batch runs the work. Other genomics teams frequently work with popular open-source languages such as WDL, Nextflow, Snakemake, and CWL. AWS also helps with this – the Amazon Genomics CLI (AGC) is free and makes it simple to deploy the necessary AWS infrastructure for your genomics workflows written in these languages. Thanks to AGC, you can use Batch queues, manage instance fleets, and even run a Nextflow pipeline on AWS without much cloud knowledge. It handles the hard work needed to configure and scale cloud resources for genomics applications. That means a bioinformatician can quickly launch an existing pipeline from nf-core on AWS, with AGC distributing the right resources among the tasks and increasing them as needed. Alternatively, bioinformatics pipelines can be run using workflow engines on AWS container services (using ECS or EKS for Nextflow’s Kubernetes executor, as an example), and the main point is that AWS has several tools to help you organize and run complex projects in the cloud.
AWS Managed Databases and Analytics
Sometimes, you must query or visualize the results after the initial analysis. Amazon Aurora and DynamoDBare AWS services that allow software programs to store and retrieve genomic annotations or results efficiently. You can directly analyze data in S3 using Amazon Athena, such as analyzing variant data in CSV/Parquet format. You can use Amazon SageMaker to develop and run ML models on your genomic data, since it provides a fully managed platform – this includes building polygenic risk score models and image classifiers for pathology slides with your AWS-based data.
Combining fast storage, highly scalable compute, and strong orchestration results in an adaptable genomics platform on AWS for any size project. Now, we will look at the stages you can follow to use your cloud genomics pipeline successfully.
Step-by-Step: Running Genomic Workloads on AWS Cloud
Switching to cloud-based bioinformatics may not seem easy, but handling it in little steps makes it much easier. The following are helpful tips and advice for running your genomic workloads on AWS:
Plan Your Cloud Genomics Architecture
You should begin by checking the specifications needed for your genomic workflows (amount of data, the number of computations, the stages in your pipeline). Choose AWS services that best fulfill your needs and arrange them in a suitable architecture. You need to determine the location of your input data (such as on S3 with FASTQs), the tool you will use for batch computing (such as AWS Batch or your own setup), and the way your pipeline will be arranged (Step Functions, Nextflow, Cromwell, etc.). Building a solution blueprint firstensures that storage, compute, workflow engine, and networking are compatible. The genomic framework from AWS helps you make a safe and adaptable system using top recommendations. Also, think about the locationof your infrastructure (typically go for a place near sequencing labs or users, while also respecting any data residency rules).
Making Product Discovery Clear and Accessible
Transform Concepts into Products in Four Weeks with Our Proven TechBoost Program
See Product Discovery ServicesEstablish Secure Data Storage and Transfer
Get your landing zone for data up and running in AWS. Usually, you need to create at least one Amazon S3 bucket to store your genomic data. Collect your data into buckets or prefixes based on whether it is raw, in process, or final, to make it easier to control access and polices. Use encryption on data you store (S3 will handle it automatically, and you can handle the encryption keys through AWS KMS). If you are sequencing by yourself, think about moving your files to S3: you can use AWS DataSync to automatically and quickly move data from your lab to S3 as soon as it is created. Thanks to DataSync, genomic data is transferred securely, at any scale, and at low cost, which is essential since it often includes protected health information (PHI). If you find internet bandwidth is slowing things down, try using AWS Snowball devices to transfer your data. Finally, establish rules that will transfer data you rarely access to less expensive storage, saving costs in the long run.
Leverage Cloud-Native Workflow Management
Instead of analyzing data manually, let a workflow engine use AWS’s capabilities to handle the scaling. If you have already set up your pipelines with WDL or Nextflow, use the Amazon Genomics CLI (AGC) to arrange your AWS resources. AGC will automatically create AWS Batch, AWS Step Functions, and Amazon CloudWatch for you, so you can run your workflows by issuing a single CLI command. According to AWS, AGC “makes it simpler and more automated to deploy the needed cloud infrastructure for genomics.” AGC is compatible with open workflows from nf-core, letting you start immediately with the best workflow. If you choose, you can submit your bioinformatics jobs to AWS Batch by building and running Docker images, or you could use a workflow manager like Nextflow to do the same. The idea is to use AWS automation fully – let the cloud handle making, running, and deleting servers, instead of treating it as a permanent group of servers. This system allows you to work more quickly and manage larger sample volumes simultaneously when needed.
Choose the Right Compute Resources (and Optimize Them)
You can pick the right computing resource for every part of your AWS pipeline. Pick an instance type based on your application. For example, genome assembly and large variant calling require memory-optimized instances, read alignment uses compute-optimized instances, and AI models or GPU-accelerated tools work best with GPU instances. With AWS Batch, you can organize your jobs using different types or sizes of instances in the queues. Set up autoscaling for your cluster so that EC2 instances are launched when jobs are added to the queue and shut off when the jobs are complete, ensuring you don’t pay for idle resources. When looking for affordability, try Spot Instances on Amazon EC2, since they can save you up to 90% compared to regular ones. Because re-trying tasks is often possible in genomic workflows, Spot is a good match. Actually, you can use our solutions to manage checkpoints and continue your work uninterrupted on Spot, without worrying about any loss. For instance, using MemVerge’s SpotSurfer, long-term jobs can be resumed when a Spot instance is taken back, thanks to transparent checkpoint/restart. It doesn’t matter if you use additional tools; design your pipeline to handle failures by using retry strategies from AWS Batch or your workflow engine to deal with Spot instances being interrupted or errors. As a result, the system provides a reliable and economical option for working with genomics.
Implement Strong Security and Compliance Controls
Since genomic data is sensitive, it must be protected, and regulatory rules must always be followed. AWS has several options to help ensure your genomics platform is compliant. To get started, put your compute instances and databases in a private Amazon VPC so they are inaccessible from the internet. Use VPC endpoints to move data to S3 without going through public networks. Apply AWS Identity and Access Management(IAM)to ensure access is limited – you might give a role permission to read or write S3 buckets or start AWS Batch jobs, but no other tasks. Enable encryption for your data both when at rest and in transit. Because AWS complies with HIPAA and GDPR, it is important to do things like sign a Business Associate Agreement for HIPAA (if necessary) and use AWS Artifact for compliance reports. Keep an eye on your environment using AWS CloudWatch and AWS CloudTrail – both tools create logs showing who accessed your data and when, which supports your compliance with data use rules. Following “security by design” right from the beginning ensures that your clinical genomics workloads on AWS are protected and private.
Experience Expert IT Consultancy
Transformative Strategies for Your Technology Needs
Discover IT ConsultingMonitor, Optimize, and Iterate
After starting genomic workloads in the cloud, get into the habit of checking and improving them regularly. Use Amazon CloudWatch to monitor your pipeline’s metrics, including how much CPU each instance uses, how long jobs take, and the rate at which data is being moved. Many genomics teams use CloudWatch metrics or logs to keep an eye on important steps in their pipelines, such as reading speed. AWS Cost Explorer and billing alarms can tell you if your process is using more storage space than intended or your pipeline is running longer than you set. Take your observations and modify your workflows: it could lead to better performance if you pick different instance types or alter how much work is done at once. AWS users should keep up-to-date with the latest AWS services and features. For example, AWS could introduce a new instance or service like Amazon Omics to help you work faster and more easily. There are always new developments in the cloud. If you update your setup occasionally, you will keep your work at the top level of efficiency and capability. Overall, cloud genomics requires ongoing updates, but eventually, both the process and outcomes will be more efficient and less costly.
Example architecture of a cloud-based genomics workflow on AWS
The Amazon Genomics CLI is used in this reference design to automate building the entire bioinformatics pipeline. All raw DNA reads are kept in Amazon S3’s input bucket. Triggering the Genomics CLI from the Jupyter Notebook on SageMaker allows the user to run a workflow written in Nextflow, WDL, and so on, on AWS Batch’s elastic compute fleet. In the workflow, every task runs in a container on the batch compute instances (including alignment and variant calling), and the intermediate results are saved to S3. The completed results are analyzed in a SageMaker notebook using R or Python for both statistics and visualization. Workflow engine, Batch queues, Spot instances, and other AWS infrastructure are all handled and scaled automatically by the Genomics CLI. This automation demonstrates that AWS removes the usual need for managing HPC clusters so that teams can concentrate on their analysis work.
For lab automation ideas, explore Automation & AI: How Genetic Testing Labs Beat Staffing Shortages.
Empowering Genomic Insights with AWS Cloud
Any organization wanting to speed up genomics progress can run their genomic tasks on the AWS cloud. With AWS’s flexible structure and many available services, small groups can access supercomputing resources whenever needed. In the cloud, bioinformatics pipelines adjust according to the demands of both the data and the question being studied. Consequently, sequencing projects are completed sooner, teams can work together internationally on common data, and researchers can try new ways of analyzing results without spending a lot upfront.
The advantages for business stakeholders are equally obvious. With cloud-based genomics on AWS, genetic research costs are now variable and depend on project activity, rather than being fixed. With just a few clicks, teams can begin a project and make changes, and if the system needs to process 10x more data in the next month, it can. In addition, AWS always brings new services (such as better HPC systems and Amazon Omics) that allow your genomics platform to take advantage of new technology without needing a significant overhaul. Genomics moves very fast, so the ability to adapt is essential.
All in all, organizations and research groups can carry out their genomics workloads properly in the cloud by starting with a thoughtful architecture, choosing appropriate AWS services, optimizing expenses and security, and turning to automation. The infrastructure created can handle the extensive DNA data of today, as well as any even larger amounts expected in the future. AWS offers the tools, and it is your job to use them to analyze genomes further. The rise of genomics will mean that anyone using cloud computing will be best placed to lead advances in precision medicine, new bioinformatics tools, and scientific discoveries. Run your bioinformatics pipelines on the cloud to elevate your genomic research.
Leading Development Teams for Your Success
Optimize Your Project Execution with Our Dedicated Development Teams
Get Your Development TeamLet’s connect and identify how we can support you in designing and building your reliable bioinformatics platform in the cloud.
For financial planning tips, refer to Lean AWS Cost Optimization: The Definitive Guide for Startups & CTOs.
Key Takeaways
- Using the cloud to handle large genomics data analysis tasks is important.
- Using AWS, you can quickly adjust your infrastructure needs, make it compliant, and keep costs low.
- The core of many bioinformatics pipelines is built on AWS Batch, EC2, and S3.
- By using containers, the app can be moved and copied easily.
- AWS speeds up discoveries in genomics and the launch of new business ideas.